The story of its development now spans from fundamental research in academia to a transformative technology to tackle targets with the potential to significantly impact patient lives. Mass spectrometry (MS) itself is almost a century old and has become an indispensable tool in many disciplines of science by allowing researchers to make extremely accurate measurements of molecular weight.
The technique analyses compounds in the gaseous phase and has hence been historically limited to small molecule analysis — during which the required vapour pressure of the analyte could be achieved at temperatures below those needed to decompose the samples.
The restrictions that confined mass spectrometry to the world of small molecule analysis were eroded through a series of scientific breakthroughs by various researchers. One of the most notable was the coupling between electrospray ionisation and mass spectrometry, first achieved by Malcolm Dole, and then later by John Fenn, for which he received the Nobel prize in chemistry in 2002.1
The technique allowed extremely large, involatile molecules (such as proteins) to be transferred to the gas phase in an ionised form via electrospray ionisation (ESI), making them suitable for MS analysis.
Once it was understood that the ESI-MS approach could preserve the non-covalent interactions that were necessary for the higher order structure of biomolecules, the field of native MS was born.2
An early advocate of these new approaches was Professor Dame Carol Robinson; having completed a PhD in the application of MS to study the structures of bioactive compounds, she became fascinated with this emerging approach and was keen to develop the technique and understand its potential to solve fundamental problems in biology.
As the new field emerged, so too did fundamental challenges regarding the ability to record the spectra of proteins. Mass spectrometers were classically designed and built to transmit small molecular weight ions. Prof. Robinson therefore formed an early collaboration with a mass spectrometry manufacturer in the UK to implement a series of instrument modifications that allowed improved transmission; focusing of large molecular weight ions, this pushed the mass/charge (m/z) window into the hundreds of thousands.
With these developments came the potential to study increasingly complex macromolecular structures, such as highly dynamic ribosomes.3 Her early work began to highlight how native mass spectrometry was able to differentiate itself from the various other biophysical approaches traditionally employed in structural biology.
The technique was particularly useful when looking at dynamic assemblies, for instance; the flexible parts of theses assemblies are difficult to capture in a crystallography experiment or multicomponent pathways with many coexisting equilibria.3,4
Native mass spectrometry in drug discovery
Despite the growing success of native MS in protein research, its application was largely driven by a small number of academic groups who possessed the necessary know-how. Ultimately, several limitations of the technique remained that inhibited its widespread adoption as a tool in a drug discovery setting — although some of the more innovative pharmaceutical companies had begun to use it.
One major hurdle, particularly for drug discovery applications, was the incompatibility of membrane proteins with the approach, which accounts for approximately 60% of all current drug targets. Membrane proteins natively reside in the hydrophobic environment of the cell membrane, which means that once isolated and purified, they are not water soluble.
Traditionally, detergents are supplemented to solutions at concentrations above their critical micelle concentration (cmc), whereby the formation of micelles around hydrophobic regions of membrane proteins allow them to be solubilised.
The heterogeneity of micelles, in terms of the number of detergent molecules present within each assembly, means that the native mass spectra acquired for these assemblies are plagued with several overlapping peaks that are extremely complex and uninterpretable.
Despite several attempts to get an intact membrane protein complex into a mass spectrometer, without the loss of its structure and interactions with neighbouring proteins, it was finally achieved in Prof. Robinson’s laboratory at Oxford University.5
The solution to the problem was uncovered by operating the instrument using unusually energetic parameters. It was realised that with the controlled application of collision energy within the mass spectrometer, micelle structures could be selectively removed before inducing excessive subunit dissociation of the protein complex of interest.
Understanding the potential of membrane proteins, as well as the common hurdles faced by other biophysical techniques in their analysis, Prof. Robinson focussed on developing these applications. Her latest work highlights the potential of introducing membrane proteins directly from membranes isolated from their native environments and being able to assess downstream signalling in real-time.6,7
A company is born
This is a tantalising prospect for drug discovery because membrane-embedded receptors and transporters are arguably the most challenging drug targets to study … but remain an extremely desirable class in diseases with high unmet need.

The increased attention from pharmaceutical companies and the intellectual property generated from Prof. Robinson’s lab provided the opportunity to form a spinout company. The new company, OMass Technologies, provided a framework to begin applying the technology to highly relevant and challenging pharmaceutical targets. Working across various drug discovery pipelines, OMass was able to understand the value of native MS.
The considerable research focus, particularly towards G-protein-coupled receptors (GPCRs), has elicited some exciting developments for the technology. By monitoring the interactions formed between GPCRs and their signal transducers (G proteins), the functional effect of a compound on the receptor could be captured.
The ability to understand which compounds invoke the desired pharmacological response in the target at the very early hit finding stages provided a unique screening platform. As a result, OMass was able to strategically pivot from a CRO to an independent drug discovery company.
The ambitious move required a significant refinancing in a series A funding round, eventually totalling £41.5 million and a complete restructuring of the company’s board, executive team and scientific expertise.
Industrialising the native MS platform
The process of transforming the fundamental technology into an engine for drug discovery required its integration with several other techniques that, together, make up a wider drug discovery platform. The close integration of native MS with deep expertise in membrane protein biochemistry is essential for the successful deployment of native MS against challenging targets, so much so that these disciplines are locked into a continuous feedback cycle.
We refer to the combined unification of these capabilities as the OdyssION platform (Figure 1), established to allow us to preserve a target’s downstream interaction partners and provide information regarding how hit compounds — from our custom-designed chemical libraries — affect the function of the target.
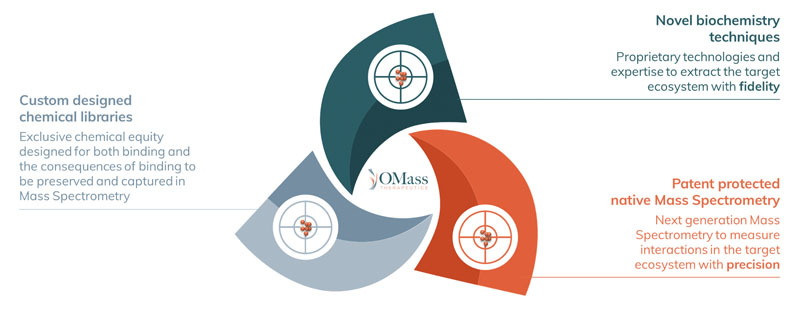
Figure 1: The OdyssION drug discovery platform
Several additional technical advantages of using native MS as a screening technique have further expanded the differentiation offered by the approach. The ability to detect weak, functional hits owing to the sensitivity of the approach enables researchers to develop chemistries and build elucidate structure activity relationships (SARs).
The platform has hence been generating promising data using fragment-based compound libraries. These benefits increase the probability of finding progressible hits and higher quality chemical starting points. In addition, the platform allows OMass to uncover new biology, leveraging natural allosteric sites and identifying binders without the bias of predetermined activity.
OMass is deploying its platform to drive drug discovery efforts against numerous targets in immunology and genetic diseases, all with the potential of delivering life-changing therapies to patients. Prof. Robinson remains a very active member of the OMass team and currently leads the company’s scientific advisory board.
References
- J.B. Fenn, “Electrospray Ionisation Mass Spectrometry: How it all Began,” J. Biomol. Tech. 18(3), 162–172 (2007).
- J.A. Loo, “Studying Noncovalent Protein Complexes by Electrospray Ionization Mass Spectrometry,” Mass Spectrom. Rev. 16(1), 1–23 (1997).
- L.L. Ilag, et al., “Heptameric (L12)6/L10 Rather than Canonical Pentameric Complexes are Found by Tandem MS of Intact Ribosomes from Thermophilic Bacteria,” Proc. Natl Acad. Sci. USA 102(23), 8192–8197 (2005).
- I-O. Ebong, et al, “Heterogeneity and Dynamics in the Assembly of the Heat Shock Protein 90 Chaperone Complexes,” Proc. Natl Acad. Sci. USA 108(44), 17939–17944 (2011).
- N.P. Barrera, et al, “Micelles Protect Membrane Complexes from Solution to Vacuum,” Science 321(5886), 243–246 (2008).
- D.S. Chorev, et al, “Protein Assemblies Ejected Directly from Native Membranes Yield Complexes for Mass Spectrometry,” Science 362(6416), 829–834 (2018).
- S. Chen, et al., “Capturing a Rhodopsin Receptor Signalling Cascade Across a Native Membrane,” Nature (2022): doi.org/10.1038/s41586-022-04547-x.
