The work is the result of a massive fragment screening effort to develop an antiviral targeting the SARS-CoV-2 main protease. The project was led by Martin Walsh, Deputy Life Sciences Director at Diamond Light Source, Frank von Delft, Professor of Structural Chemical Biology at the University of Oxford and Principal Beamline Scientist of I04-1/XChem at Diamond, and Nir London, Assistant Professor at the Weizmann Institute Israel.
The team combined mass spectrometry with the XChem facility at Diamond, the UK’s national synchrotron, to rapidly identify new lead compounds for drug development to treat COVID-19.
For this study, called crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease, the team probed an essential enzyme of SARS-CoV-2 with more than 1250 unique small compound, termed fragments, and identified 74 high-value fragment hits that can be used to develop new inhibitors for this essential viral protein.
The paper details the data along with proposed design routes for progressing towards improved, more potent, compounds.
“COVID-19, caused by SARS-CoV-2, lacks effective therapeutics. Additionally, no antiviral drugs or vaccines were developed against the closely related coronavirus, SARS-CoV-1 or MERS-CoV, despite previous zoonotic outbreaks."
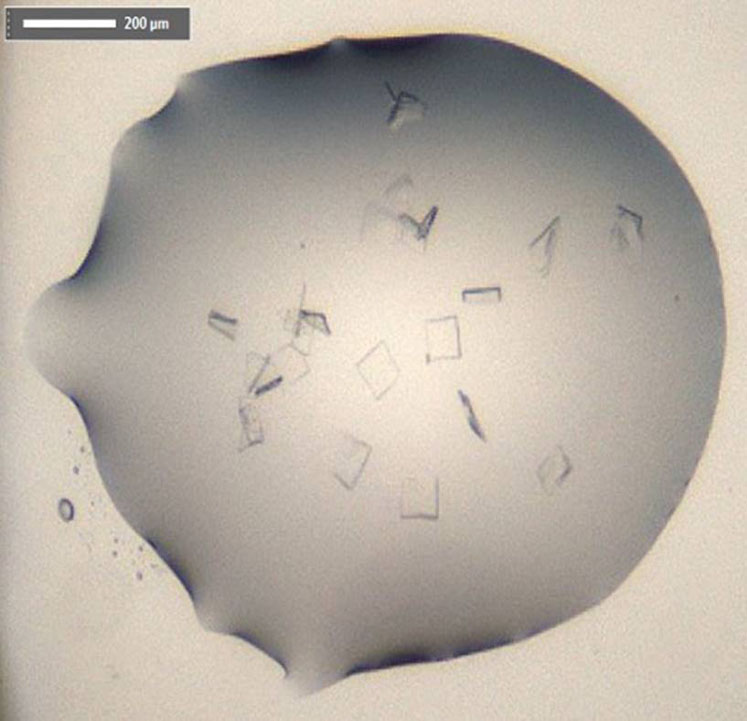
Structure of SARS-CoV-2 main protease: crystals of CoVID-19 free enzyme
"To identify starting points for such therapeutics, we performed a large-scale screen of electrophile and non-covalent fragments through a combined mass spectrometry and X-ray approach against the SARS-CoV-2 main protease, one of two cysteine viral proteases essential for viral replication."
"Our crystallographic screen identified 74 hits that span the entire active site, as well as three hits at the dimer interface. These structures reveal routes to rapidly develop more potent inhibitors and offer unprecedented structural and reactivity information for on-going structure-based drug design against SARS-CoV-2 main protease,” explains Martin Walsh who, in addition to his role at Diamond, is also a Medical Research Council (MRC) funded Research Group Leader at the Research Complex at Harwell (RCaH).
Structural biology, which can play a key role in drug development, was also rapidly deployed after the 2002 SARS-CoV-1 outbreak, with earlier work by the Hilgenfeld group on the main protease of coronaviruses leading to crystal structures of SARS-CoV-1 protease and inhibitor complexes.
Other studies have taken the popular approach of a high-throughput screens (HTS) using very large compound libraries, followed by structural studies to elucidate the binding mode.
“Despite these efforts, drugs remain elusive that directly target SARS-CoV-2 (rather than disease symptoms) and are verified by clinical trials. In retrospect, this is perhaps unsurprising for the main protease inhibitors, as both peptidomimetic and covalent inhibition carry risks as strategies for drug development."
"In general, the simpler the molecule the lower the risk. We, therefore, applied a different approach to this protease, using fragment screening by high-throughput structural biology,” adds Martin Walsh.
Fragment methods have become a staple of modern drug discovery, using small collections (100 s or 1000 s) of small compounds (<300 Da) that bind promiscuously and thus sample a far larger chemical space than is achieved by HTS.
The challenge is that the very weak binding of fragment hits requires highly sensitive biophysical detection, careful confirmation of binding and specialised medicinal chemistry expertise take the hits and develop them into fully potent drug candidates.
However, the real promise of fragments with the right expertise and equipment, they can quickly and efficiently be converted into valid drug candidates with a much simpler route to clinical impact.
Rapid advances in technology and automation at synchrotron radiation sources has made screening directly in crystal structures routinely possible at facilities such as the XChem platform at Diamond Light Source.
The team took the highly unusual route of releasing all the experimental data as soon as it was generated; the announcement on social media triggered a large international collaboration that harnessed the combined knowledge of scientists worldwide through a novel crowdsourcing initiative that they called COVID Moonshot.
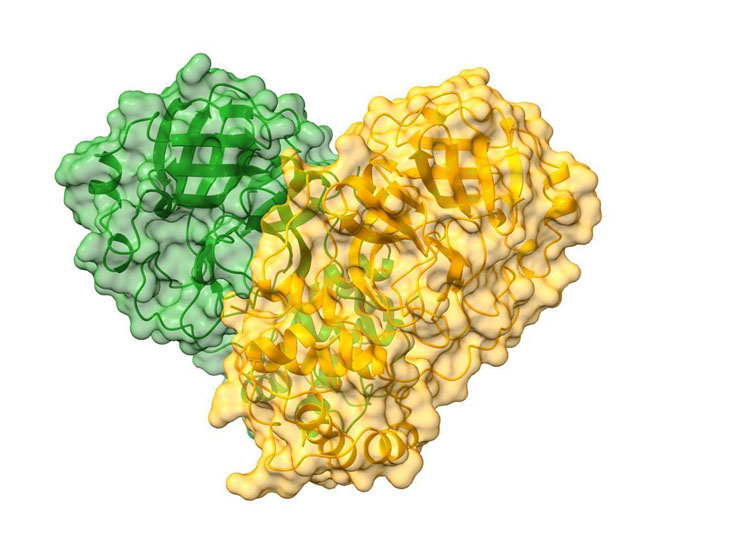
Cartoon representation of the CoVID-19 dimer with a semi-transparent surface in green and orange delineating each monomer
“Performing the experiment and achieving the high data quality in a few weeks, as lockdown started, was a tour de force and a credit to our highly talented scientists. Even more remarkable was the response of the international community to the data release: it mobilised a vast pool of expertise, technologies and philanthropy, which evolved into a unique and rigorous drug discovery effort that aims to develop rapidly an entirely novel, easily synthesised, oral antiviral with good safety and pre-clinical properties."
"Working fully in the open, data are released near real-time, so the outcome will be available to any drugs manufacturer worldwide. The world’s focus has been on vaccines and repurposing of existing drugs, but the Moonshot is one of a small number of projects attempting novel small molecule therapeutics,” said Frank von Delft.
SARS-CoV-2 is a large enveloped, positive-sense, single stranded RNA betacoronavirus. The viral RNA encodes two open reading frames that, through ribosome frame-shifting, generates two polyproteins pp1a and pp1ab.
These polyproteins produce most of the proteins of the replicase-transcriptase complex. The polyproteins are processed by two viral cysteine proteases: a papain-like protease (PLpro), which cleaves three sites, releasing non-structural proteins nsp1-3 and a 3C-like protease, also referred to as the main protease (Mpro), that cleaves at 11 sites to release non-structural proteins (nsp4-16).
These non-structural proteins form the replicase complex responsible for replication and transcription of the viral genome and have led to Mpro and PLPro being the primary targets for antiviral drug development.
